Thomas Greatrix, Frank C. Langbein, Roger M. Whitaker, Gualtiero B. Colombo, Liam D. Turner. High-Confidence Labelling of Pathology Reports using LLM-Based Unanimous Ensembles with Limited Data. Proc AI in Healthcare (AIiH), Cambridge, UK, September 2025. [PDF]
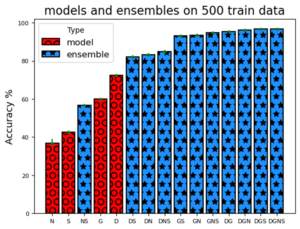
Manual labelling of pathology reports is a costly bottleneck for medical data analysis. We propose diverse unanimous ensembles, integrating Large Language Models (LLMs) like GPT-4o with complementary model architectures, for high-confidence automatic labelling of pathology reports, particularly addressing the challenge of labelled training data scarcity. This consensus method yields high precision on an automatically identifiable subset while simultaneously flagging ambiguous cases requiring expert review. Applying this to the public TCGA-Reports dataset, a GPT-4o and DistilBERT ensemble achieved 95.5% accuracy on the 45.5% subset representing a 23.1 percentage point increase over the baseline DistilBERT’s overall accuracy on the full dataset. This demonstrates potential for cost-effective data annotation by automatically labelling high-confidence subsets, thereby reserving human effort for ambiguous cases.
![]() This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.